A pandoc-based layout workflow for scholarly journals
23 Mar 2018In my work, an academic publishing service for Open Access journals, we never provided any solution for layout and typesetting of articles. Journals with no support in this complex task explored a variety of solutions, mostly dependent on the most common routines from authors (and of course from editors too) in their field of studies. The two main options are the use of LaTeX, in the few happy disciplines whose authors are accustomed with this typesetting technology, and the use of external professional support — mostly based on InDesign. These two options are naturally restricted to certain disciplines or to the (rare!) availability of funds. Thus for the majority of our Editorial Teams the only available option has been the use of Microsoft Word and other word processors.
There is a number of limits in these solutions, let’s face the main one: the only publication format promptly available is the PDF, the de facto standard for scientific articles which mimics the old times of paper, while any future-proof format should require at least a non-fixed layout and some semantic expressiveness.1 For these solutions, the preparation of articles in HTML format is non-straightforward at best, and often nearly impossible in a consistent way and with good quality. There are also other — sometimes very serious — limits on the quality of the PDF, with common problems such as: broken urls, lack of an index and metadata, and (mostly when a word processor was involved) awful typesetting and some drama due to rasterized texts. For Editorial Teams working with word processors, this also implied a lot of wasted time with the formatting, micromanaging of spaces and styles, and an uncomfortably high amount of mistakes: something better and smarter was badly needed.
In 2017 I finally looked for a nice solution for the preparation of multiple publication formats. The following is still an experimental workflow and it is meant to be a good solution for scholarly journals that use OJS for submission process and publication. It is based on the awesome pandoc and relies on the pairing of markdown (for full-text) and YAML (for metadata and settings), as a more friendly and simple alternative to XML.
See markdown-workflow in Github
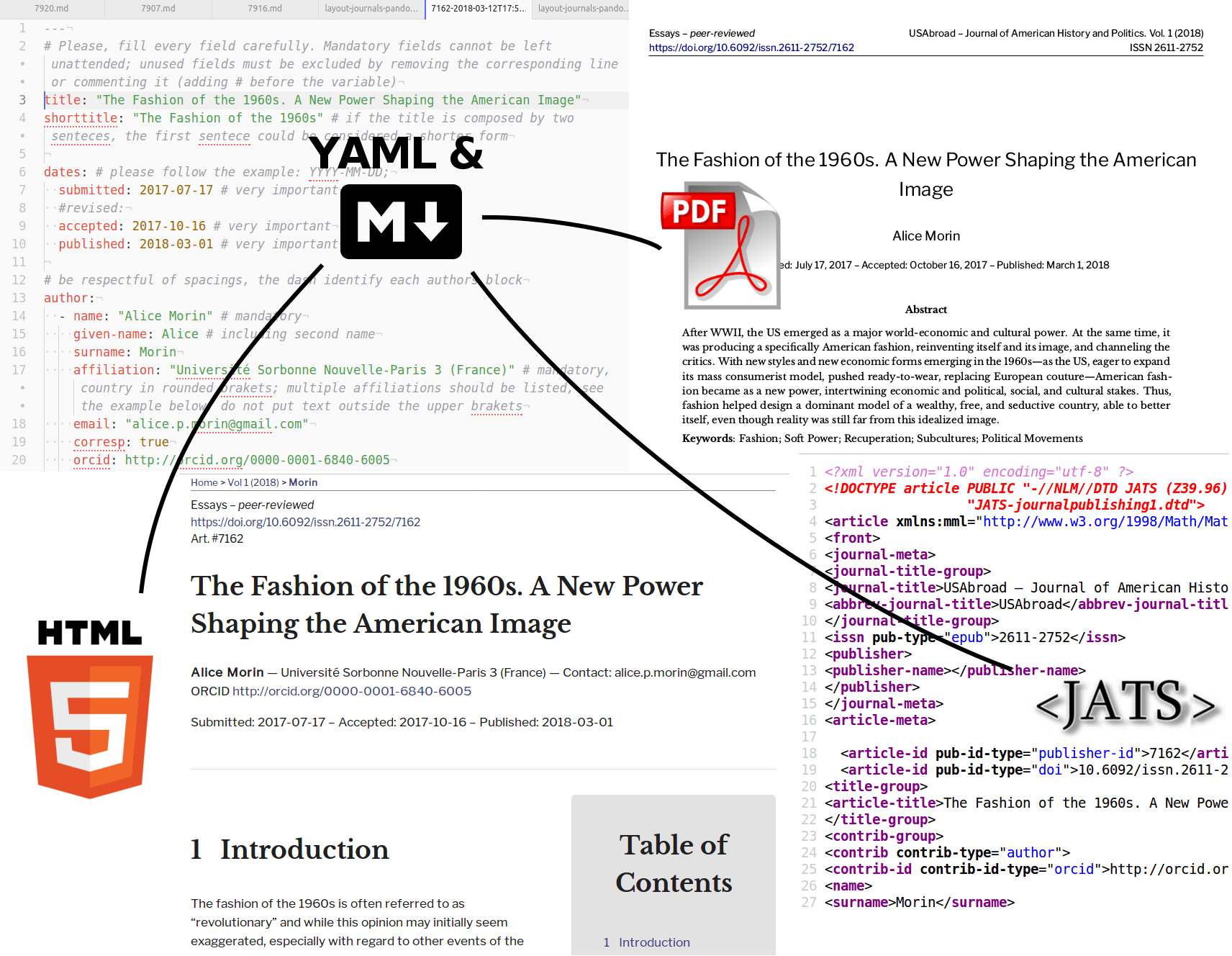
Two formats to rule them all
Why another solution?
NB: Writing these notes, I stumbled upon a recently published paper that covers the same problems following basically this same path, while being more evolved:
Albert Krewinkel and Robert Winkler, «Formatting Open Science: agilely creating multiple document formats for academic manuscripts with Pandoc Scholar», PeerJ Computer Science 3 (8 May 2017): e112; https://doi.org/10.7717/peerj-cs.112.
There are several solutions for managing HTML scholarly articles in smarter ways, like the various iterations of Scholarly HTML and Scholarly Markdown.2 Probably the most complete and capable solution is Research Articles in Simplified HTML (RASH) Framework, that provides a complete semantic annotation for metadata, full-text and references.
Unfortunately all this projects are intended for authors rather than Editorial Teams, thus failing — in my humble opinion — to be a sustainable solution for small journals. In fact, small scholarly journals cannot require authors to sustain strict and complex requirements for paper submissions. The strong need of low complexity for article and metadata syntaxes (regarding editors), and for scripts and templates (regarding myself) are the reason I have chosen pandoc, markdown and YAML as the most practical and affordable path.
Pandoc, markdown and YAML
I already wrote a couple of posts on the power of pandoc and the simplicity of markdown and YAML.3 In short, pandoc is a powerful and universal text converter, that is able to handle the conversion of texts in a wide variety of formats.
Markdown and YAML are two light and simple4 languages that could work as an exchange format for formatted texts (markdown) and structured data (YAML). Compared to XML they are far more human readable, simple and straightforward, and their wide adoption makes them a good solution for workflows, format transitions and future reusability.
The main idea is to obtain, for each manuscript, a markdown copy to manage the “layout,” using pandoc to convert (#1) full-texts from manuscripts in DOCX or ODT5 while also extracting as much semantic expression as possible — such as headings for article sections, long quotations, footnotes, external URLs. The articles converted to markdown would also lose all the unnecessary complexity and confusion deriving from styles, custom formatting, comments, revisions etc.
After that, markdown articles could be used by the Editorial Team for the final formatting, and adjusting the markdown syntax where needed: markdown will allow them to focus on the meaning, rather than appearance, while all the graphics will be treated separately and in a more consistent way. A second task for Editorial Teams would also be the embedding for each article of its metadata in YAML: this syntax could be used inside the same markdown file, thus having a single file for full-text and metadata. Having metadata structured explicitly rather than written as normal text will allow not only for a better and consistent handling of the final graphic layout, but also for high reusability and format transition, just like a dedicated XML.
When an article in markdown + YAML is ready, pandoc can be used to convert (#2) it in multiple publication formats, provided that adequate templates are set to use the various metadata and settings inside the YAML.
The workflow
It is already a gamble to ask Editorial Teams — from Social Sciences and Humanities — to work with markdown, YAML, and a text editor, and I am happy to say that for now I won this gamble.
In order to lower the amount of complexity and to benefit from serialization, the workflow happens inside a shared folder and is governed by scripts. The shared folder permits a distributed workload, while exempting Editorial Teams from the most technical part: the use of command line tools and the setup of a LaTeX environment. This part is currently made centrally by myself, having a shared working directory for each Journal and launching manually the conversion scripts whenever necessary.
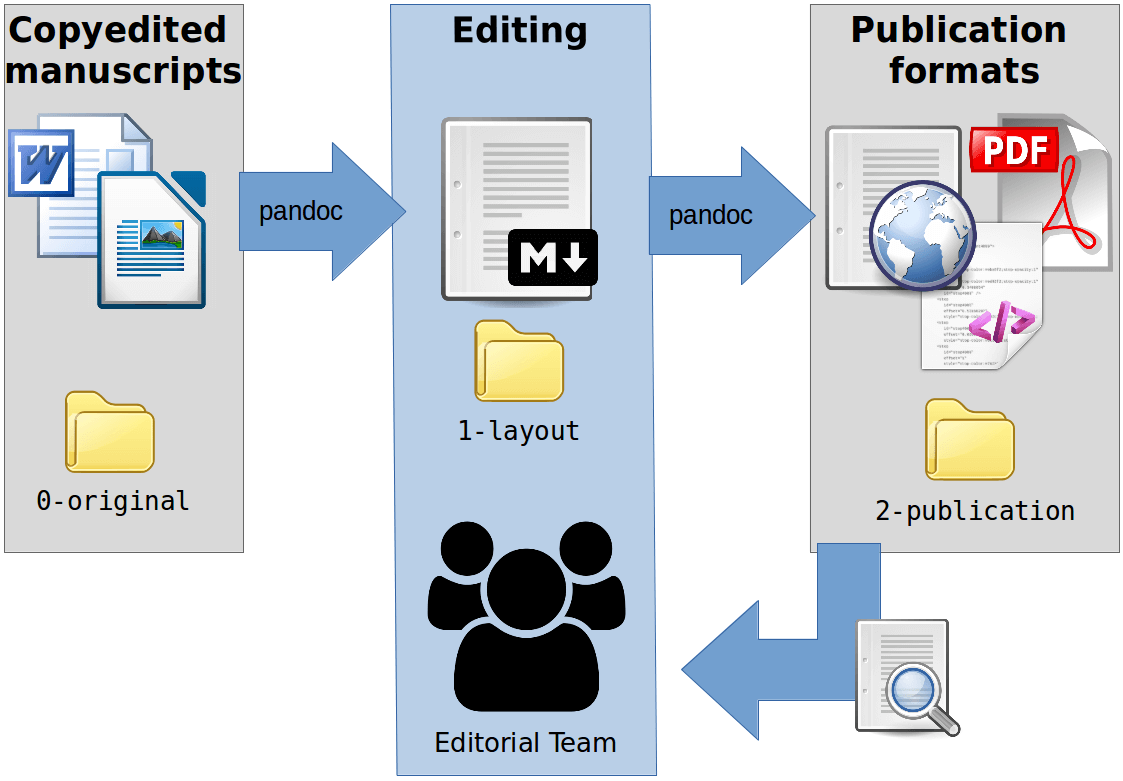
An ugly representation of the workflow
As seen in the previous section, the conversions are 2:
-
from the author’s original manuscript (peer-reviewed) to markdown:
articles in DOCX or ODT should be deposited in the appropriate directory (./0-original/) for conversion; they will be renamed with safer names (the OJS submission ID will be used if the manuscript is originated from the OJS submission process) and converted in markdown, with a default metadata section; the new converted files will be copied to the layout directory (./1-layout/) where editors can start their work with formatting and metadata; -
from markdown to the publication formats (galley files):
where markdown files in layout directory are converted in various publication formats (they will be generated in./2-publication/). This step is meant to be repeated until the articles will be ready for publication.
Each conversion is scripted to be applied to all the available articles within the source directories, and it includes the creation of backup copies.
Two more scripts are intended for basic image processing (images are resized, if needed, and a lower resolution for HTML is created) and for archiving the working directory after the issue is published. The latter will also provide a “self-contained” version of markdown manuscripts, in which metadata and configurations at both journal- and issue-level are appended.
Publication formats
HTML is the native format of World Wide Web, and as such should be the main option for scholarly communications too. HTML for article publication gives some considerable advantages:
- an easier readability, regardless of the devices;
- advanced semantic annotations;
- a nice format for digital preservation (the files are also self-contained, with images embedded in base64).
Semantics was added taking examples from previous (and more advanced) experiences with scientific HTML, such as scholarly HTML and RASH. HTML articles are annotated with Schema.org, starting from ScholarlyArticle property, using RDFa Lite. Unfortunately a proper annotation of the body of the paper has not been made yet, thus the RDFa Lite is applied only to paper metadata.
The PDF will be generated by pandoc via XeLaTeX engine, thus allowing for UTF-8 support and custom fonts for each Journal, aside for the excellent typesetting of LaTeX. The template uses hyperref for linked citations and allow to use up to three different fonts, for full-text, titles, and paratext.
JATS XML and TEI XML are two critical formats for achieving interoperability with other platforms; output in these two formats is possible, though the templates need further testing.
Published issues
Three journals has published an issue with this workflow until now, while three more journals should join the first group of early adopters:
- Cinergie, No 12 (2017): the first Editorial Team to help me testing this workflow; the issue consists of 28 items (1 editorial, 23 articles and 4 reviews), all published in HTML and PDF; 11 papers contain images, although the possibility to handle two different file versions for PDF and HTML was not ready at the time. PDF are typeset in Libre Caslon Text and Lato (for titles and paratext).6
- ZoneModa Journal, Vol 7 (2017): the issue consists of 18 items (2 editorials, 8 articles and 8 non-refereed items), plus an “intermission” edited separately; 5 items contain images, in two different formats for PDF (in a resolution up to 300DPI) and HTML (low resolution). Article sections are unnumbered. PDF are typeset in EB Garamond, a custom font for titles and Ubuntu font for paratext.6
- USAbroad, Vol 1 (2018): a (still open) issue that consists of 1 editorial and 5 articles, with indented text; this was also the first issue that undergone the full OJS submission workflow. PDF are typeset in Libre Baskerville and Libre Franklin (for titles and paratext).6
All the issues include also an (unpublished) JATS XML version for papers.
By the way all the three journals adopt my standard layout for OJS 2.4 journals. The HTML stylesheet is based on the same style, it includes minimal rules embedded in the HTML file (table of contents, margins, indent rules), while the most part of the style is taken from the journal stylesheet. Some parts, such as abstracts and author’s biography, are hidden when viewed within the journal website, since they are redundant in that context.
Limitations and known issues
Currently this solution is still rudimentary and requires some dumb cut&paste to import metadata from OJS that should be possible to automatize, and doesn’t exploit the full potentiality of pandoc. Further improvements could come from the use of filters, i.e. for linked references to images and tables, and for more flexibility regarding metadata.
Time permitting, I would also explore the possibility to prepare a single file for issue, with table of contents and colophon, in PDF and ePub.
This workflow was completed notwithstanding the small amount of dedicated time and my very limited experience in scripting, LaTeX, semantic annotations etc., thus I will warmly appreciate every feedback, suggestion, and help.
-
although we are still quite far from the goals of Tim Berners-Lee & James Hendler in this short (closed access!) commentary:
«Publishing on the semantic web», Nature 410, 1023–1024 (26 April 2001) https://doi.org/10.1038/35074206. ↩︎ -
Scholarly HTML, Scholarly Markdown, Academic Markdown: several projects that revived several times, for the same goal, making writing scholarly articles more smart. ↩︎
-
the two posts are currently available in Italian, but you can find a ton of better resources in English around the web. ↩︎
-
“simple” here is to be intended as the contrary of “complex,” not as a synonym for “poor.” ↩︎
-
I would love to try to convert from other formats, such as LaTeX. ↩︎
-
A special mention for these beautiful open fonts: Libre Caslon Text, Libre Baskerville and Libre Franklin are from Pablo Impallari (et. al.); EB Garamond is a gorgeous Garamond from Georg Mayr-Duffner, it is also a very complete font with robust support of Greek and Cyrillic scripts. ↩︎ ↩︎ ↩︎
comments powered by Disqus